Photoreactions are chemical reactions occurring from an electronic excited state produced by light absorption, to another electronic state named the photo-product. Molecular photophysics describes photoreactions as electronic transitions between molecular states represented by potential energy surfaces (PES). Many photoreactions occur through conical intersections (CInt), which are specific molecular geometries where distinct PES intersect. At such a CInt, the electronic transition between the photo-excited state and the photoproduct is controlled by the nuclear motions resulting from the specific PES topography (shape), itself strongly influenced by the interactions of the molecule with its environment in the condensed phase.
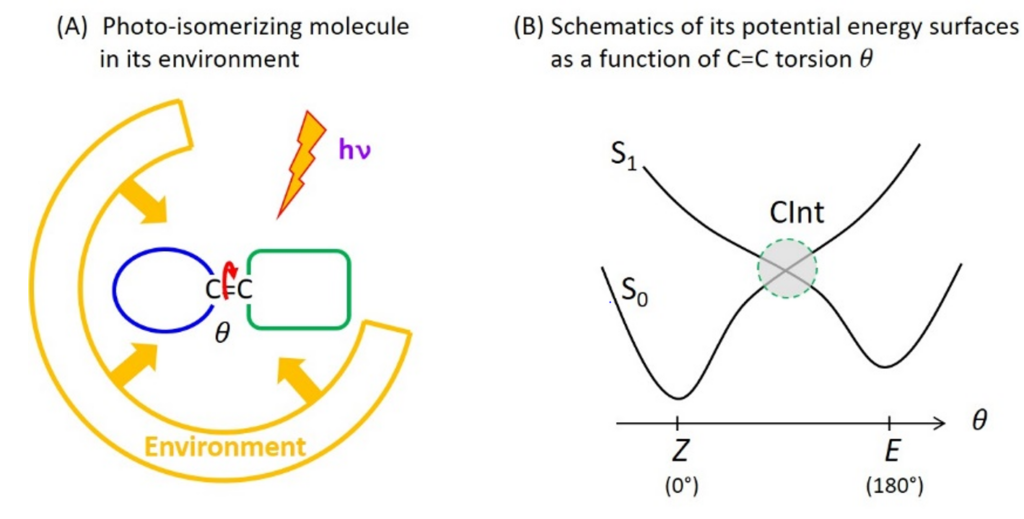
C=C double bond photoisomerisation is an example of photoreaction occurring through a CInt (figure 1). In particular, the photoisomerisation of retinal in the protein for vision (rhodopsin) has long been a prototype photoreaction – studied both experimentally by ultrafast spectroscopy and theoretically by quantum chemistry – for the fundamental investigation of the physico-chemical factors that control the speed (i.e. excited state lifetime) and efficiency (i.e. quantum yield) of photoreactions occurring through a CInt.
In the BioDyn team we contribute to such investigations along two directions: (i) controlling (i.e. increasing) the excited state lifetime – hence fluorescence quantum yield – of microbial retinal proteins for optogenetic applications, or (ii) improving the photoisomerisation quantum yield in molecular photoswitches or photo-motors for the conversion of light energy into mechanical energy at the molecular scale.
Retinal proteins
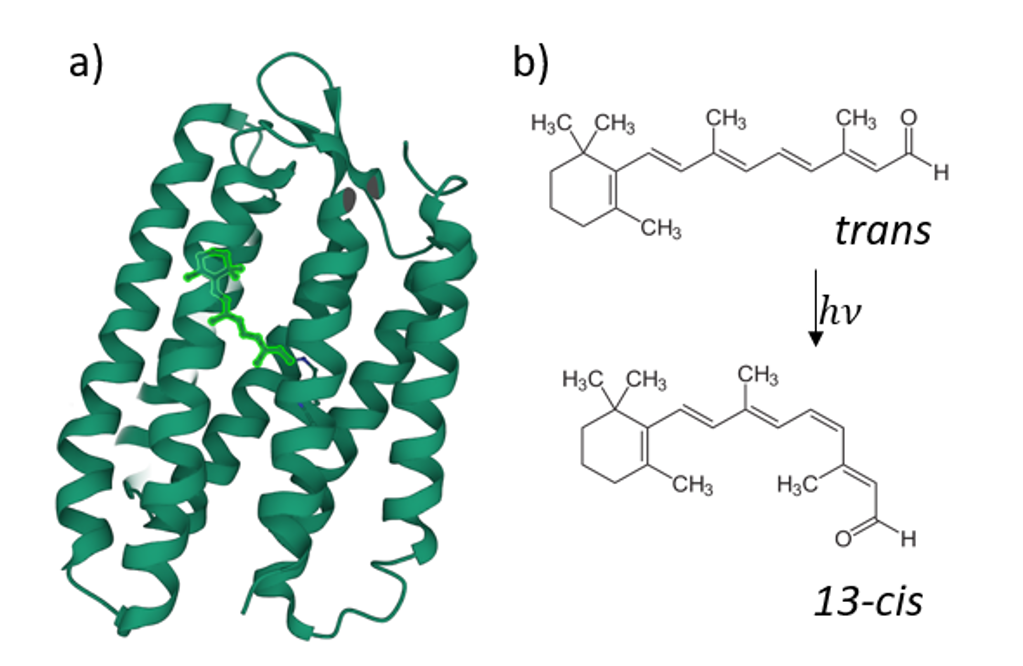
Retinal proteins use light for a large variety of bio-molecular processes. The most prominent member of this family is rhodopsin, the visual pigment of all vertebrates.
It was recently found that the fluorescence quantum yield (FQY) of the reaction of retinal all-trans/13-cis -photoisomerisation has a strong dependence on the transmembrane voltage for some retinal proteins (figure 2), such as Archaerhodopsin-3 [1]. This ability was then put forward as a possible candidate for opto-genetic investigations, i.e. in the form of a genetically encoded voltage indicator (GEVI) [2] to track neuronal electric signals [3]. However, higher FQY would be required for in vivo applications. As it was shown before, protein mutation has such an effect [4]. That’s why an understanding of the mechanistic role of the mutations that enhance the low fluorescence signal of the mutants is crucial for designing more efficient rhodopsin-based fluorescent reporters.
What does ultrafast spectroscopy of retinal proteins provide in this context?
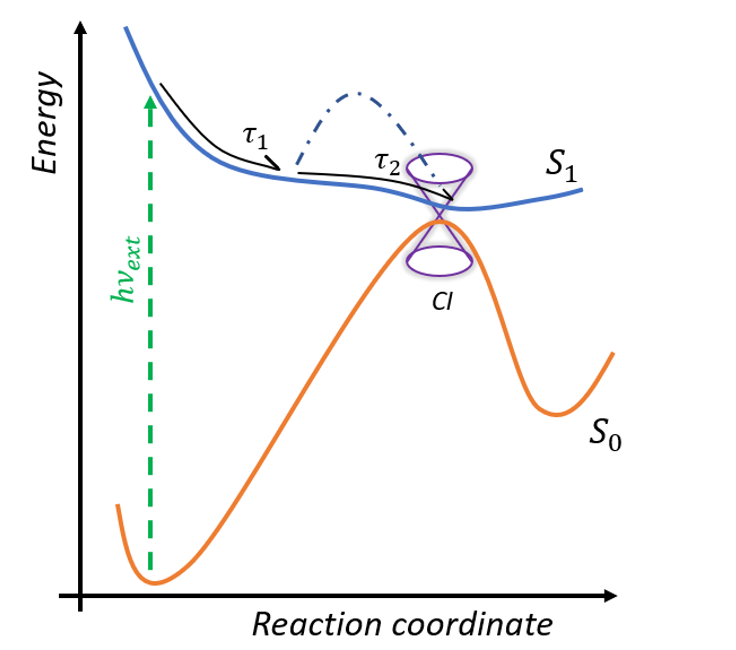
The photoreaction inside retinal proteins occurs at the sub-picosecond time scale. After excitation from the ground S0 to the first excited state S1, the system relaxes from Franck -Condon geometry to the minimum of the excited state in the sub-picosecond range and then dependent on the form of the potential surface goes to the conical intersection (figure 3) [5]. The time of this progression determines the FQY of the proteins. After the CInt, the system relaxes to the photoproduct (cis-isomer) or back to the initial configuration. Ultrafast fluorescence and transient absorption spectroscopy in the UV/VIS/near IR spectral domain are clarifying the excited state dynamics and the effects of selected site mutations on the formation of the potential barrier in the excited state.
Once the precise mechanism of the enhanced FQY, i.e. prolonged excited state lifetimes, is established in terms of retinal-protein interactions, the long-term objective of this project is to design fluorescent mutants of AR-3 for applications in optogenetics. This work is a part of ANR project UltraArchae in collaboration with scientists from France (LASIR), UK, Italy and Japan.
Bio-mimetic photo-switches and molecular motors
In a C=C double bond photoisomerisation reaction, the torsion motion leads to the conversion of light energy to mechanical energy (rotational movement). This motion can be back and forth torsion as in the case of molecular switches [6], or can be unidirectional as in the case of Light driven molecular motors (LDMM) [7] .
We are intrigued by the following challenges: How to engineer – i.e. design chemically – a molecular motor/ switch in order to control motion at the molecular scale with an optimum efficiency – i.e. photoisomerisation Quantum Yield (QY)? One strategy to address these questions is to investigate and possibly mimic the very efficient C=C double bond photoisomerisation of retinal in the rhodopsin protein (the pigment for vision). Hence, we investigate so-called biomimetic molecular switches or motors to unravel how the potential energy surfaces (PES) and the molecular environment (in condensed phase) control the vibrational motions and eventually the QY. Can we discover rules or recipes for the chemical design of molecular structures producing the desired PES shapes?
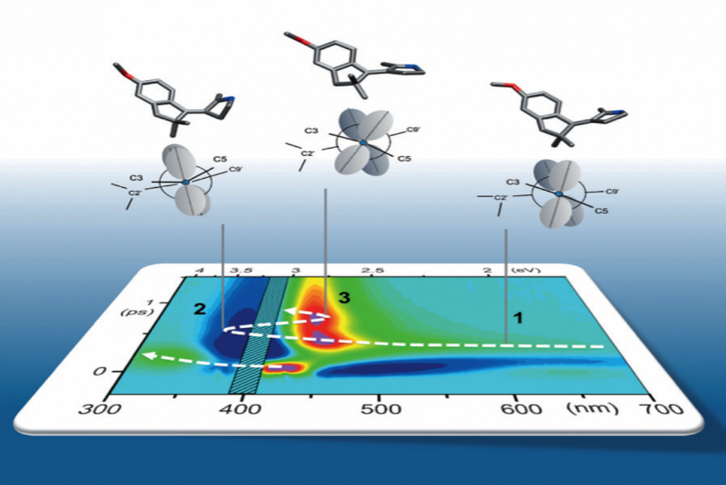
We implement Transient Absorption spectroscopy (TAS) to monitor the evolution of the spectroscopic signatures of molecules while they evolve from the initial Franck-Condon state on the first excited state (S1) towards the CInt, and beyond after decay to the ground state (S0), see figure 4. Such observations are carefully analyzed and compared to theoretical predictions made by quantum chemical simulations performed by various collaborators. In this context, one experimental challenge is to monitor the signatures of vibrational motions along the photoisomerisation reaction. This can be done by performing vibrational coherence spectroscopy using a sub 10-fs excitation pulse in a dedicated TAS experiment. [8]
References:
[1] D. Maclaurin, V. Venkatachalam, H. Lee, A.E. Cohen (2013). Mechanism of Voltage-sensitive fluorescence in microbial rhodopsin. Proceedings of the National Academy of Sciences, 110.15. 5939–44. https://doi.org/10.1073/pnas.1215595110
[2] Y. Bando, C. Grimm, V. H. Cornejo, R. Yutse. (2019). Genetic voltage Indicator. https://doi.org/10.1186/s12915-019-0682-0
[3] J. M. Kraji, A. D. Douglass, D. R. Hochbaum, D. Maclaurin, A. E. Cohen (2011). Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nature Methods,90–95 https://doi.org/10.1038/nmeth.1782.
[4] R. S. McIsaac M.K.M Engqvist, T. Wannier et al (2014). Directed evolution of a far-red fluorescent rhodopsin. Proceedings of the National Academy of Sciences. 13034–39 https://doi.org/10.1073/pnas.1413987111
[5] C. Chang, H. Kuramochi, m. Singh, R.Abe-Yoshizumi, T. Tsukuda, H. Kandori, T. Tahara (2021). A Unified View on Varied Ultrafast Dynamics of the Primary Process in Microbial Rhodopsins Angewandte Chemie International Edition. https://doi.org/10.1002/anie.202111930.
[6] M. Paolino, M. Gueye, E. Pieri, M. Manathunga, S. Fusi, A. Cappelli, L. Latterini, D. Pannacci, M. Filatov, J. Léonard, and M. Olivucci (2016). Design, Synthesis, and Dynamics of a Green Fluorescent Protein Fluorophore Mimic with an Ultrafast Switching Function. JACS. https://pubs.acs.org/doi/pdf/10.1021/jacs.5b10812
[7] M. Filatov(Gulak) M. Paolino, R. Pierron, A.Cappelli, G. Giorgi, J. Léonard, M. Huix-Rotllant, N. Ferré, X. Yang, D. Kaliakin, A. B Gonzalez, and M. Olivucci (2022).Towards the engineering of a photon-only two-stroke rotary molecular motor. Nature communications. https://doi.org/10.1038/s41467-022-33695-x
[8] M. Gueye, M. Manathunga, D. Agathangelou, Y. Orozco, M. Paolino, S. Fusi, S. Haacke, M. Olivucci, and J. Léonard (2018). « Engineering the vibrational coherence of vision into a synthetic molecular device ». Nature Communications. https://doi.org/10.1038/s41467-017-02668-w.
