- Theoretical studies by first principles and hybrid quantum mechanics / molecular mechanics (QM/MM) approaches, especially when combined with free energy surface sampling techniques such as the metadynamics, have disclosed in the last ten years new possibilities to investigate complex biochemical reactions often not directly accessible to experiments.
- Within this domain, these simulations play the role of a virtual laboratory where experiments so-called in silico become a complement and sometimes an alternative to experiments in vitro and in vivo
- In this specific field, the research lines pursued in our group the following representative studies have been conducted:
LeuRS Synthetase: Protein-RNA interaction
Starting from the crystallographic structure of the Leucyl-tRNA (LeuRS) synthethase (Nature Struct. & Mol. Biol. 2005, 12, 915), combined hybrid QM/MM and free energy sampling dynamical approach have been used to inspect the enzymatic activity of the Leucyl-tRNA for the editing of the genetic information stored into the nucleic acid. The reaction is triggered by the dissociation of a water molecule in proximity of a chemical bond (-C*-O*-) constituting the link between tRNA and the Leucyl protein. A crucial promoter of the reaction is the 3’-OH group of the cognate tRNA bound to the editing site of Leucyl, which forms a stable hydrogen bond with this peculiar catalytic water molecule. We could identify two possible reaction mechanisms for the editing reaction: In one case the 3’-OH’group of the cognate tRNA acts as a Lewis acid, and one of the protons of the catalytic water molecule becomes a temporarily shared proton between 3’-OH and this specific H2O. In the second case, the 3’-OH group of the tRNA drives, via its hydrogen bond, the catalytic water molecule towards the –C*-O*- bond and the unoccupied LUMO state, located on top of this bond, becomes the electron acceptor for the dissociating water molecule. This promotes the reaction towards the same final product of the former pathway, without involving transient proton transfers to 3’-OH.
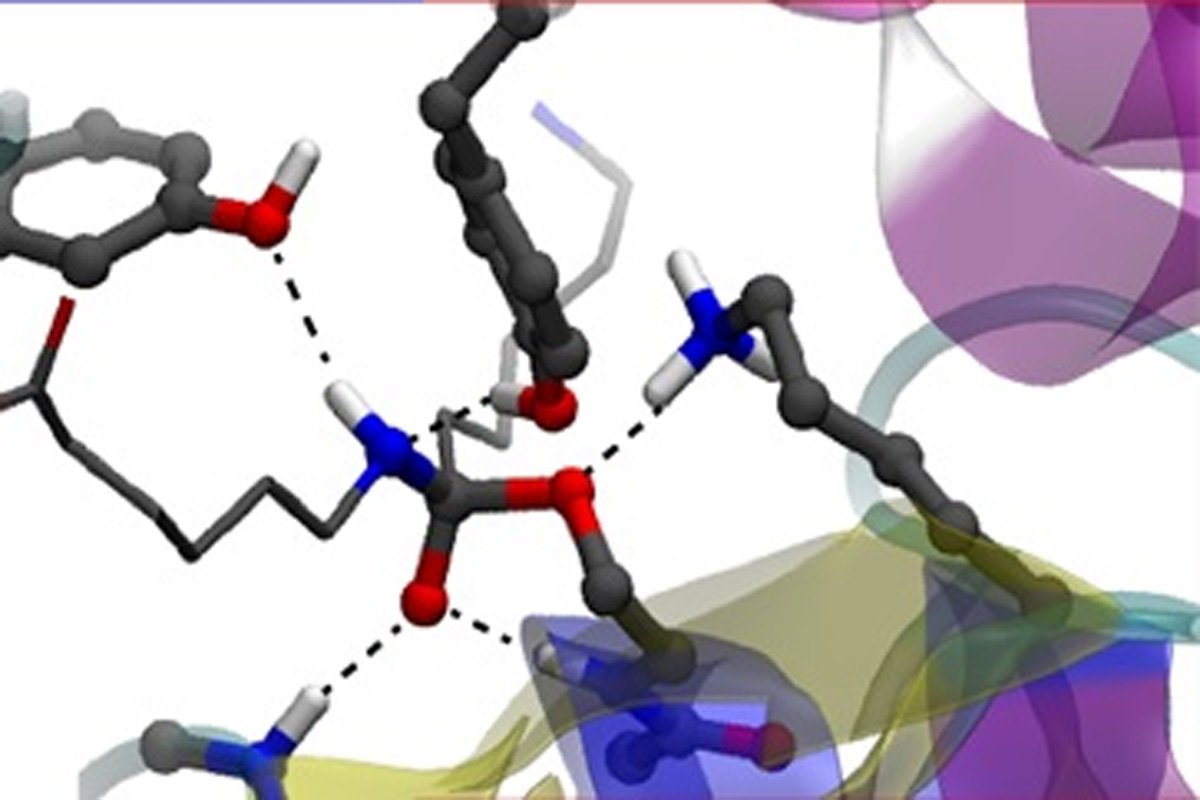
Enzymatic reactions: Degrading artificial compounds in an environmentally friendly way
The oligomers degrading enzyme Hyb-24DN has an uncommon ability to degrade artificial compounds such as nylon-6 and to promote the synthesis of biocompatibles materials. We evidenced the crucial role of a unique Tyr residue in this peculiar enzymatic reaction. The active site possesses a Ser112/Lys115/Tyr215 catalytic triad similar to the one of penicillin-recognizing family of serine-reactive hydrolases, but includes a peculiar Tyr170 residue. By using a reactive QM/MM approach, we evidenced its catalytic mechanism and related functional/structural specificities. At variance with other peptidases, we show that the involvement of Tyr170 in the enzyme-substrate interactions is responsible for a structural variation in the substrate-binding state. The acylation via a tetrahedral intermediate is the rate-limiting step driven by the catalytic triad Ser112, Lys115, and Tyr215, acting as a nucleophile, general base, and general acid, respectively. The functional interaction of Tyr170 with this triad leads to an efficient disruption of the tetrahedral intermediate, promoting a conformational change of the substrate favorable for proton donation from the general acid.

Relevant literature:
- M. Boero, J. Kang, S. Tokumoto and M. Tateno, J. Comput. Theor. Nanosci. 6, 2648 (2009)
- K. Kamiya, M. Boero, K. Shiraishi, A. Oshiyama and Y. Shigeta, J. Phys. Chem. B. 114, 6567 (2010)
- M. Boero, J. Phys. Chem. B 115, 12276 (2011)
- V. Rojas-Cervellera, A. Ardèvol, M. Boero, A. Planas, C. Rovira, Chem. Eur. J. 19, 14018 (2013)
- K. Kamiya, T. Baba, M. Boero, T. Matsui, S. Negoro, Y. Shigeta, J. Phys. Chem. Lett. 5, 1210 (2014)
- M. Boero, Reactive Simulations for Biochemical Processes, in Atomic-Scale Modeling of Nanosystems and Nanostructured Materials – Lect. Notes Phys. 795, pag. 81-98, ed. by C. Massobrio, H. Bulou, C. Goyonex, Springer, Berlin Heidelberg 2010. ISBN 978-3-642-04650-6